Comparision: FEM vs. other methods¶
In this section, we compare the performance of Free Energy Minimization (FEM) method with other existing methods based on mean field approximations and Maximum Likelihood Estimation (MLE). We shall show that FEM is more accurate than other methods. Additionally, FEM is computationally much faster than MLE.
First of all, we import the necessary packages to the jupyter notebook:
In [1]:
import numpy as np
import sys
import timeit
import matplotlib.pyplot as plt
import simulate
import inference
%matplotlib inline
np.random.seed(1)
We generate a true interaction matrix w0 and time series data
s0.
In [2]:
# parameter setting:
n0 = 40 # number of variables
g = 4.0 # interaction variability parameter
w0 = np.random.normal(0.0,g/np.sqrt(n0),size=(n0,n0))
# generating time-series data
l = int(4*(n0**2))
s0 = simulate.generate_data(w0,l)
Suppose only a subset s of variables is observed.
In [3]:
nh0 = 15
n = n0 - nh0
s = s0[:,:n].copy()
Suppose first that we know the number of hidden variables so we set
nh to its actual value.
In [4]:
nh = nh0
Let us write a plot function to compare actual couplings and inferred couplings.
In [5]:
def plot_result(w0,w):
plt.figure(figsize=(13.2,3.2))
plt.subplot2grid((1,4),(0,0))
plt.title('observed to observed')
plt.plot([-2.5,2.5],[-2.5,2.5],'r--')
plt.scatter(w0[:n,:n],w[:n,:n])
plt.xticks([-2,0,2])
plt.yticks([-2,0,2])
plt.xlabel('actual interactions')
plt.ylabel('inferred interactions')
plt.subplot2grid((1,4),(0,1))
plt.title('hidden to observed')
plt.plot([-2.5,2.5],[-2.5,2.5],'r--')
plt.scatter(w0[:n,n:],w[:n,n:])
plt.xticks([-2,0,2])
plt.yticks([-2,0,2])
plt.xlabel('actual interactions')
plt.ylabel('inferred interactions')
plt.subplot2grid((1,4),(0,2))
plt.title('observed to hidden')
plt.plot([-2.5,2.5],[-2.5,2.5],'r--')
plt.xticks([-2,0,2])
plt.yticks([-2,0,2])
plt.scatter(w0[n:,:n],w[n:,:n])
plt.xlabel('actual interactions')
plt.ylabel('inferred interactions')
plt.subplot2grid((1,4),(0,3))
plt.title('hidden to hidden')
plt.plot([-2.5,2.5],[-2.5,2.5],'r--')
plt.scatter(w0[n:,n:],w[n:,n:])
plt.xticks([-2,0,2])
plt.yticks([-2,0,2])
plt.xlabel('actual interactions')
plt.ylabel('inferred interactions')
plt.tight_layout(h_pad=1, w_pad=1.5)
plt.show()
Naive Mean-Field approximation¶
In [6]:
print('nMF:')
cost_obs,w,sh = inference.infer_hidden(s,nh,method='nmf')
w,sh = inference.hidden_coordinate(w0,s0,w,sh)
plot_result(w0,w)
nMF:
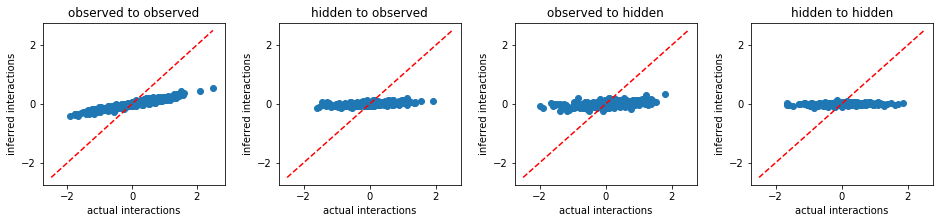
Thouless-Anderson-Palmer mean field approximation¶
In [7]:
print('TAP:')
cost_obs,w,sh = inference.infer_hidden(s,nh,method='tap')
w,sh = inference.hidden_coordinate(w0,s0,w,sh)
plot_result(w0,w)
TAP:
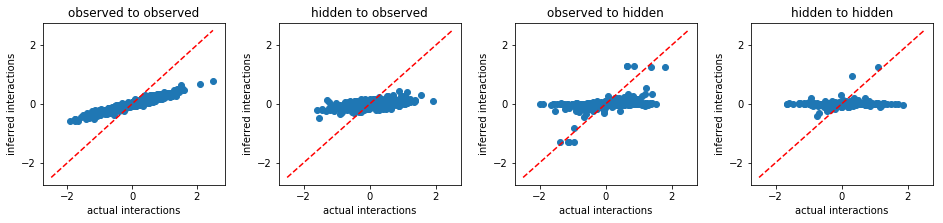
Exact mean field approximation¶
In [8]:
print('eMF:')
cost_obs,w,sh = inference.infer_hidden(s,nh,method='emf')
w,sh = inference.hidden_coordinate(w0,s0,w,sh)
plot_result(w0,w)
eMF:
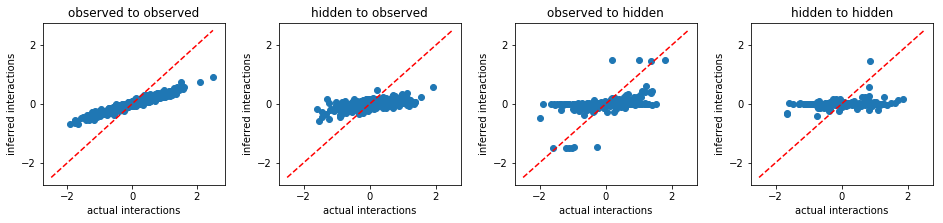
Maximum Likelihood Estimation¶
In [9]:
print('MLE:')
start_time = timeit.default_timer()
cost_obs,w,sh = inference.infer_hidden(s,nh,method='mle')
w,sh = inference.hidden_coordinate(w0,s0,w,sh)
stop_time=timeit.default_timer()
run_time=stop_time-start_time
print('run_time:',run_time)
plot_result(w0,w)
MLE:
('run_time:', 9970.889276981354)
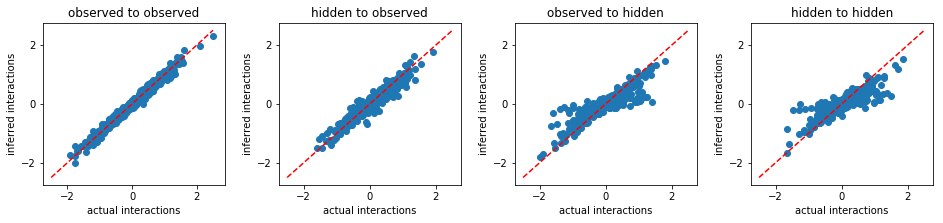
Free Energy Minimization¶
In [10]:
print('FEM:')
start_time = timeit.default_timer()
cost_obs,w,sh = inference.infer_hidden(s,nh,method='fem')
w,sh = inference.hidden_coordinate(w0,s0,w,sh)
stop_time=timeit.default_timer()
run_time=stop_time-start_time
print('run_time:',run_time)
plot_result(w0,w)
FEM:
('run_time:', 515.2393028736115)
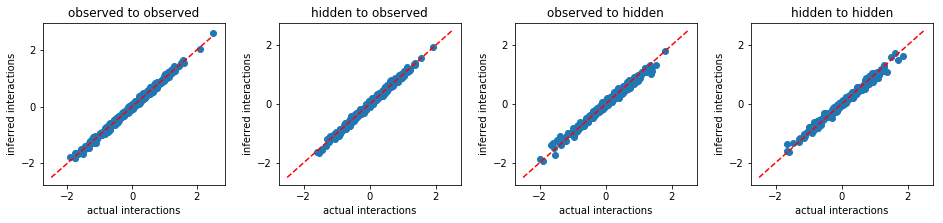
From the above results, we conclude that FEM outperforms other existing methods. Beside more accurate inference, FEM works much faster than MLE. For instance, to solve the same problem, FEM took only 515 seconds, however, MLE took 9970 seconds.
In [ ]: