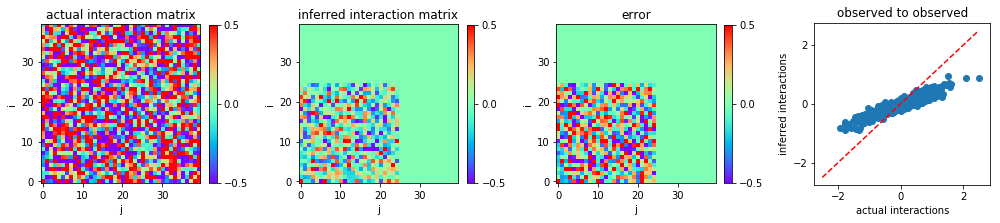Results¶
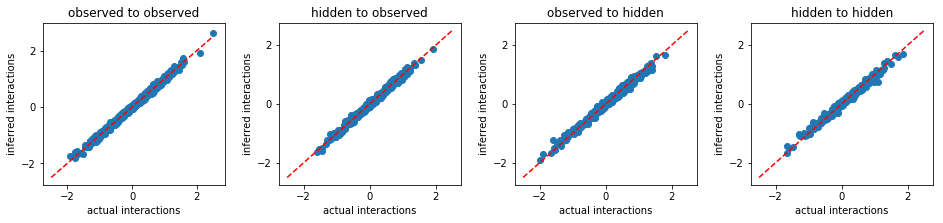
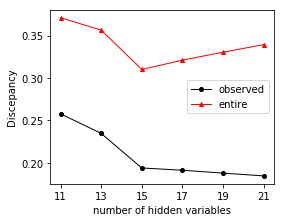
We demonstrate the performance of our method in analyzing binary data generated from the kinetic Ising model simulations in the following. Using configurations of a subset of variables, we will recover the interactions (including observed to observed, hidden to observed, observed to hidden, and hidden to hidden), the configurations of hidden variables, and the number of hidden variables.
First of all, we import the necessary packages to the jupyter notebook:
In [1]:
import numpy as np
import sys
import matplotlib.pyplot as plt
import simulate
import inference
%matplotlib inline
np.random.seed(1)
We consider a system of n0 variables interacting each others with
interaction variability parameter g.
In [2]:
# parameter setting:
n0 = 40 # number of variables
g = 4.0 # interaction variability parameter
w0 = np.random.normal(0.0,g/np.sqrt(n0),size=(n0,n0))
Using the function simulate.generate_data, we then generate a time
series of variable states according to the kinetic Ising model with a
data length l.
In [3]:
l = int(4*(n0**2))
s0 = simulate.generate_data(w0,l)
A raster of variable configurations s0 is plotted:
In [4]:
plt.figure(figsize=(8,6))
plt.title('raster of time series')
plt.imshow(s0.T[:,:400],cmap='gray',origin='lower')
plt.xlabel('time')
plt.ylabel('variable index')
plt.show()

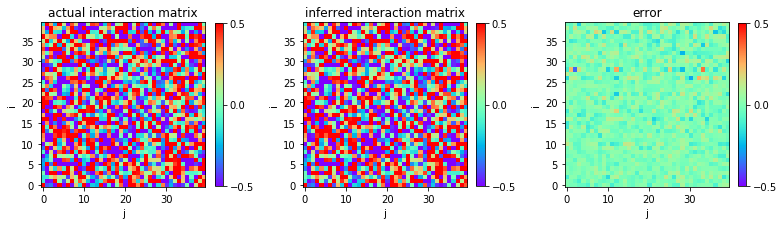
Using the configurations of entire system s0, we can infer
sucessfully the interactions between variables:
In [5]:
w = inference.er(s0)
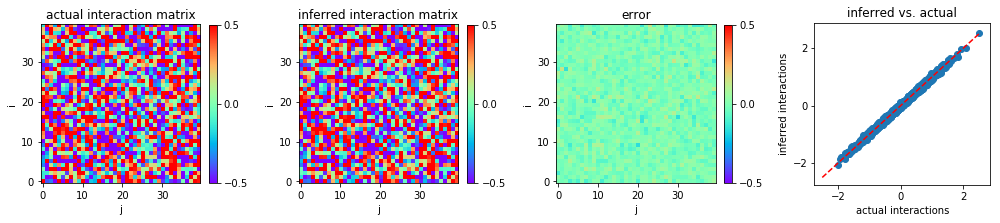
We plot the heat map of the actual interaction matrix w0, the
inferred interaction matrix w, and the error in inference, i.e., the
discrepancy between w0 and w.
In [6]:
plt.figure(figsize=(14,3.2))
plt.subplot2grid((1,4),(0,0))
plt.title('actual interaction matrix')
plt.imshow(w0,cmap='rainbow',origin='lower')
plt.xlabel('j')
plt.ylabel('i')
plt.clim(-0.5,0.5)
plt.colorbar(fraction=0.045, pad=0.05,ticks=[-0.5,0,0.5])
plt.subplot2grid((1,4),(0,1))
plt.title('inferred interaction matrix')
plt.imshow(w,cmap='rainbow',origin='lower')
plt.xlabel('j')
plt.ylabel('i')
plt.clim(-0.5,0.5)
plt.colorbar(fraction=0.045, pad=0.05,ticks=[-0.5,0,0.5])
plt.subplot2grid((1,4),(0,2))
plt.title('error')
plt.imshow(w0-w,cmap='rainbow',origin='lower')
plt.xlabel('j')
plt.ylabel('i')
plt.clim(-0.5,0.5)
plt.colorbar(fraction=0.045, pad=0.05,ticks=[-0.5,0,0.5])
plt.subplot2grid((1,4),(0,3))
plt.title('inferred vs. actual')
plt.plot([-2.5,2.5],[-2.5,2.5],'r--')
plt.scatter(w0,w)
plt.xticks([-2,0,2])
plt.yticks([-2,0,2])
plt.xlabel('actual interactions')
plt.ylabel('inferred interactions')
plt.tight_layout(h_pad=1, w_pad=1.5)
plt.show()
However, in the real world, it is likely that we do not observe the
entire system, but only a subset of variables. For instance, suppose
that there are nh0 hidden variables, so the number of observed
variables is n = n0 - nh0 and the observed configurations are
s = s0[:,:n].
In [7]:
nh0 = 15
n = n0 - nh0
s = s0[:,:n].copy()